Only available on the Enterprise planThis feature is only available on the Enterprise plan. Review our plans and pricing or sign up for our free expert-led trial today.
No required format for data files
There is no required format for data files. You can upload data in any format, whether it’s CSV, Parquet, or a proprietary data format.Parquet is a columnar storage format that is optimized for reading and writing large datasets. It is particularly useful for data that is stored in S3 buckets, as it can be read in parallel and is highly compressed.1. Prerequisites
You’ll need:- The Edge Impulse CLI.
- If you receive any warnings that’s fine. Run
edge-impulse-blocksafterwards to verify that the CLI was installed correctly.
- If you receive any warnings that’s fine. Run
- PPG-DaLiA CSV files: Download files like
ACC.csv,HR.csv,EDA.csv, etc., which contain sensor data.
- Docker desktop installed on your machine.
2. Building your first transformation block
To build a transformation block open a command prompt or terminal window, create a new folder, and run:Dockerfile
We’re building a Python based transformation block. The Dockerfile describes our base image (Python 3.7.5), our dependencies (in requirements.txt) and which script to run (transform.py).
WORKDIR under /home! The /home path will be mounted in by Edge Impulse, making your files inaccessible.
2.2 - requirements.txt
This file describes the dependencies for the block. We’ll be using pandas and pyarrow to parse the Parquet file, and numpy to do some calculations.
transform.py
This file includes the actual application. Transformation blocks are invoked with three parameters (as command line arguments):
--in-fileor--in-directory- A file (if the block operates on a file), or a directory (if the block operates on a data item) from the organizational dataset. In this case theunified_data.parquetfile.--out-directory- Directory to write files to.--hmac-key- You can use this HMAC key to sign the output files. This is not used in this tutorial.--metadata- Key/value pairs containing the metadata for the data item, plus additional metadata about the data item in thedataItemInfokey. E.g.:
{ "subject": "AAA001", "ei_check": "1", "dataItemInfo": { "id": 101, "dataset": "Human Activity 2022", "bucketName": "edge-impulse-tutorial", "bucketPath": "janjongboom/human_activity/AAA001/", "created": "2022-03-07T09:20:59.772Z", "totalFileCount": 14, "totalFileSize": 6347421 } }
unified_data.parquet is in the same directory):
out/ directory containing the RMS of the X, Y and Z axes. If you inspect the content of the file (e.g. using parquet-tools) you’ll see the output:
If you don’t have parquet-tools installed, you can install it via:
3. Pushing the transformation block to Edge Impulse
With the block ready we can push it to your organization. Open a command prompt or terminal window, navigate to the folder you created earlier, and run:
The transformation block in Edge Impulse
edge-impulse-blocks push and the block will be updated.
4. Uploading unified_data.parquet to Edge Impulse
Next, upload theunified_data.parquet file, by going to Data > Add data… > Add data item, setting name as ‘Gestures’, dataset to ‘Transform tutorial’, and selecting the Parquet file.
This makes the unified_data.parquet file available from the Data page.
5. Starting the transformation
With the Parquet file in Edge Impulse and the transformation block configured you can now create a new job. Go to Data, and select the Parquet file by setting the filter todataset = 'Transform tutorial'.
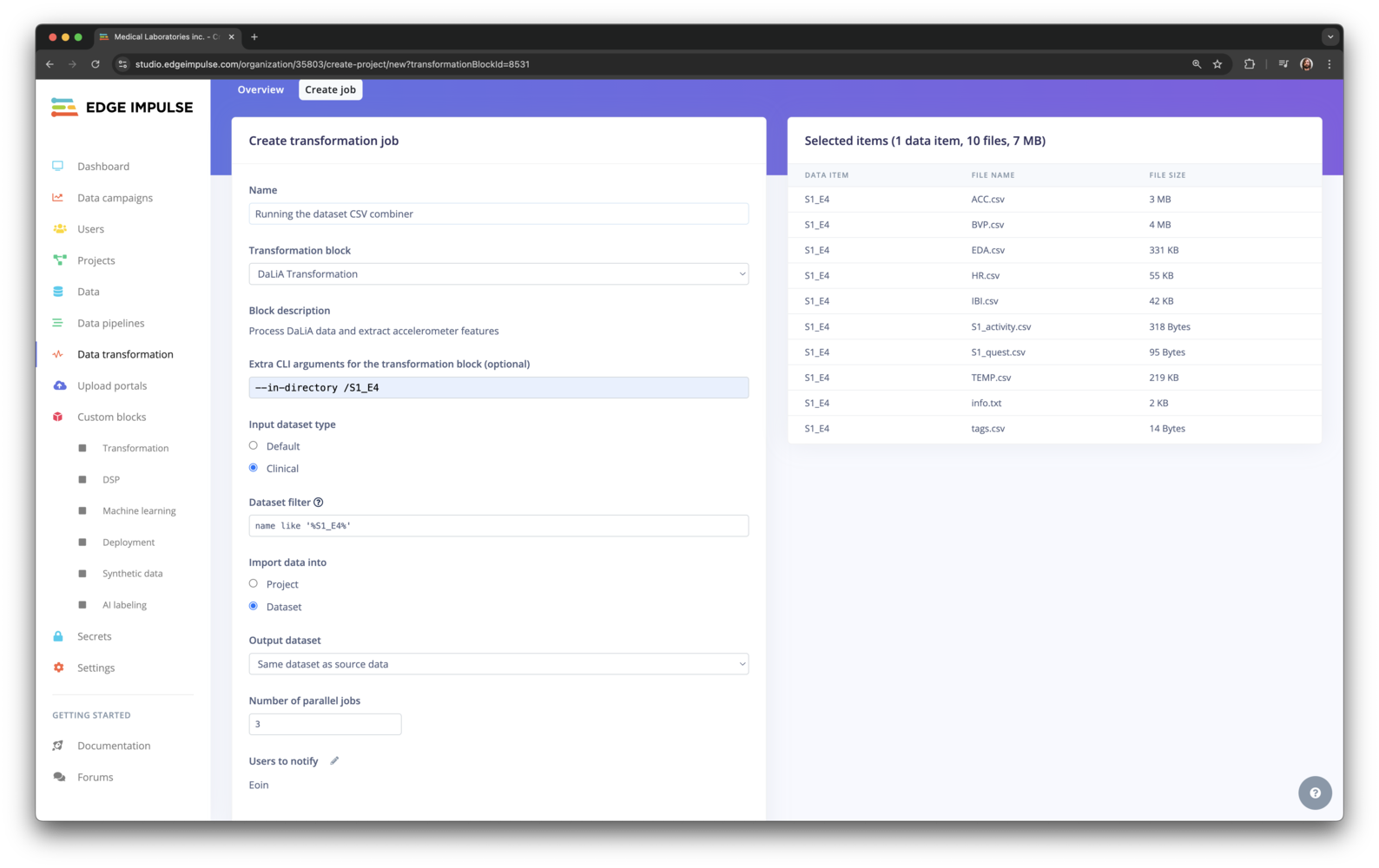
Selecting the transform tutorial dataset
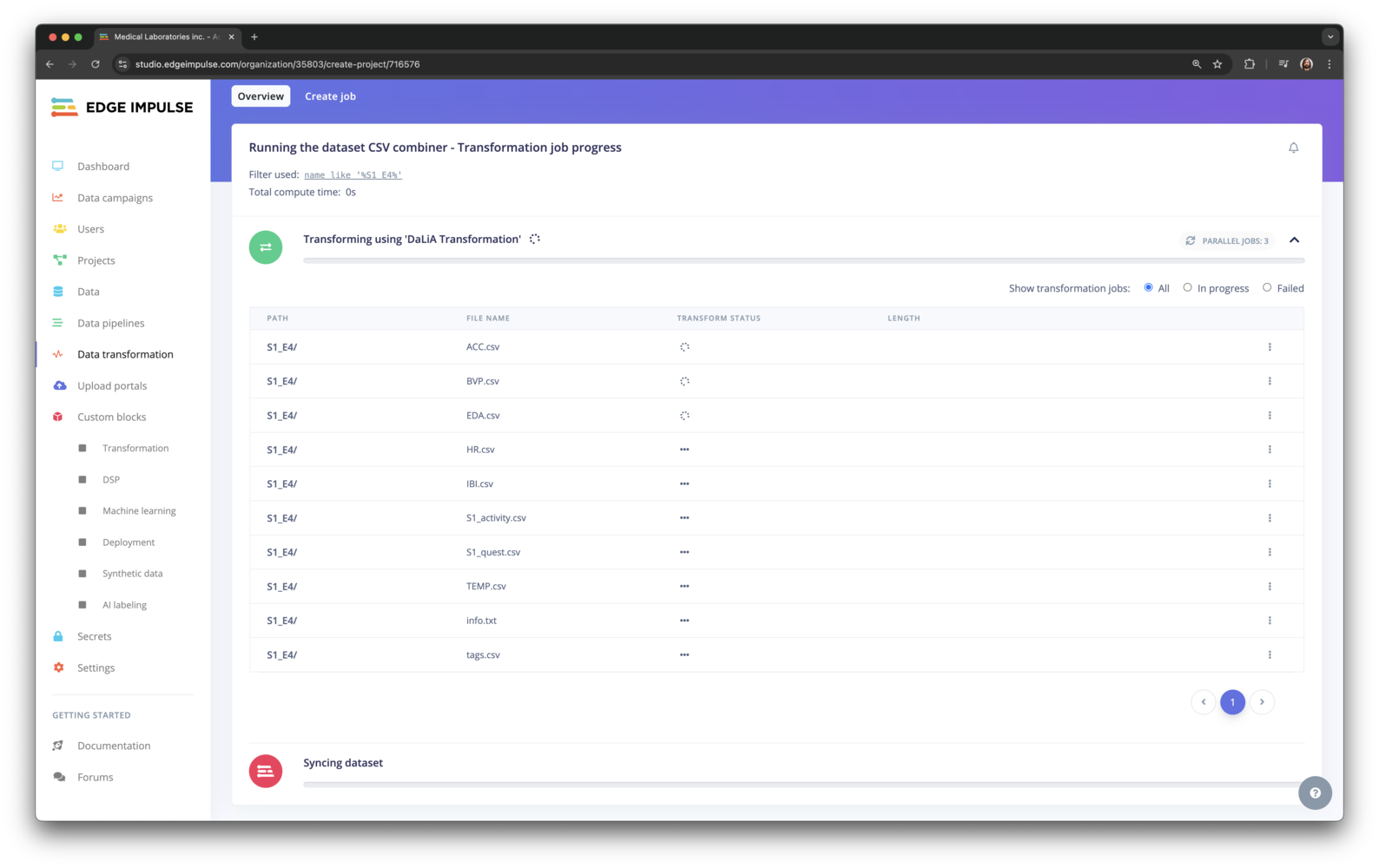
Configuring the transformation job
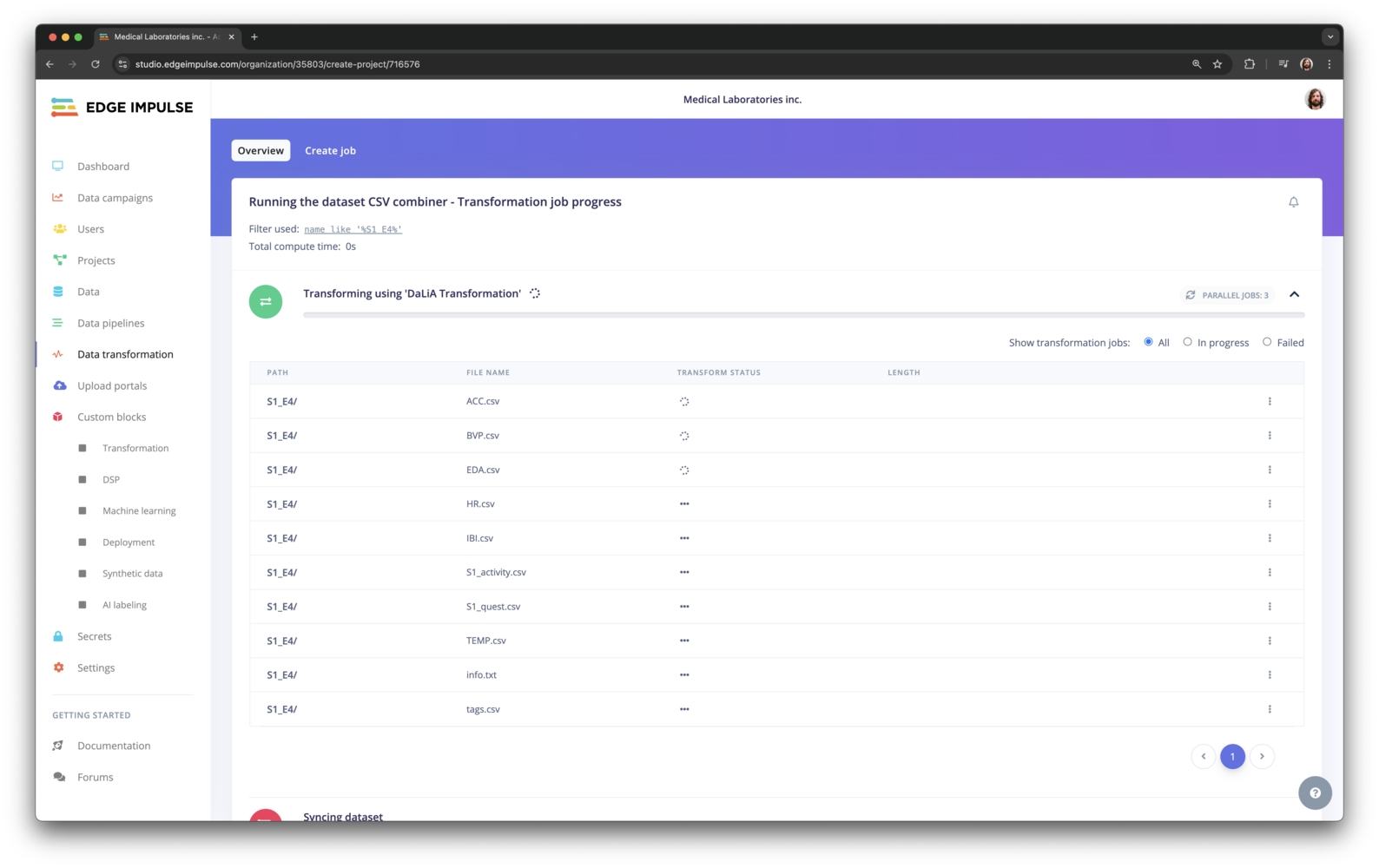
Dataset transformation running
6. Next steps
Transformation blocks are a powerful feature which let you set up a data pipeline to turn raw data into actionable machine learning features. It also gives you a reproducible way of transforming many files at once, and is programmable through the Edge Impulse API so you can automatically convert new incoming data. If you’re interested in transformation blocks or any of the other enterprise features, let us know! :rocket:Appendix: Advanced features
Updating metadata from a transformation block You can update the metadata of blocks directly from a transformation block by creating aei-metadata.json file in the output directory. The metadata is then applied to the new data item automatically when the transform job finishes. The ei-metadata.json file has the following structure:
- If
actionis set toaddthe metadata keys are added to the data item. Ifactionis set toreplaceall existing metadata keys are removed.
EI_API_KEY- an API key with ‘member’ privileges for the organization.EI_ORGANIZATION_ID- the organization ID that the block runs in.EI_API_ENDPOINT- the API endpoint (default: https://studio.edgeimpulse.com/v1).
.png?fit=max&auto=format&n=x9Ga-7v4NxdQ7jXX&q=85&s=122f826b6c39dbd7bfeb9bcbffecd095)
Running a transformation block with custom parameters