Only available on the Enterprise planThis feature is only available on the Enterprise plan. Review our plans and pricing or sign up for our free expert-led trial today.
Overview
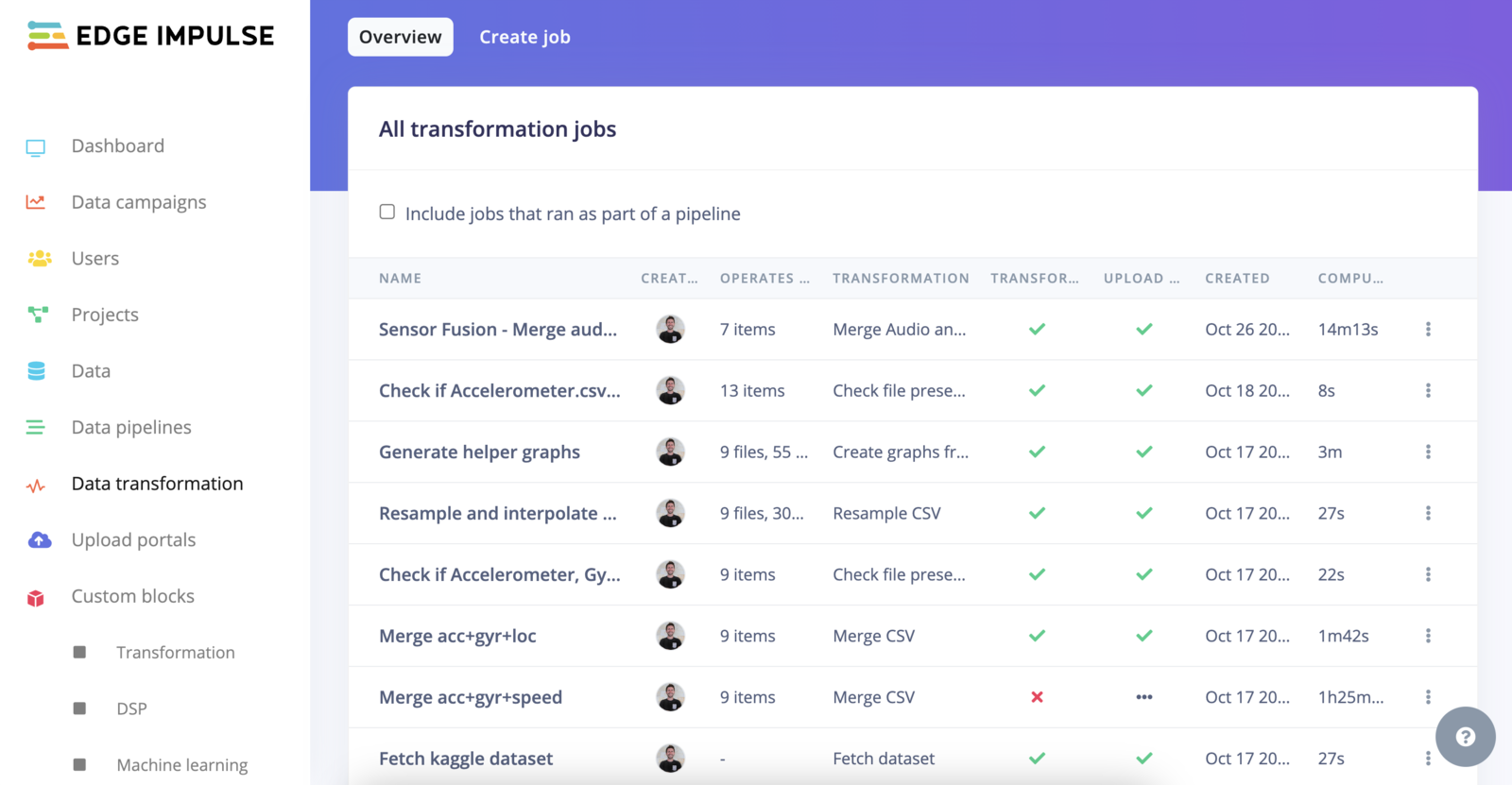
Transformation jobs
Create a transformation job
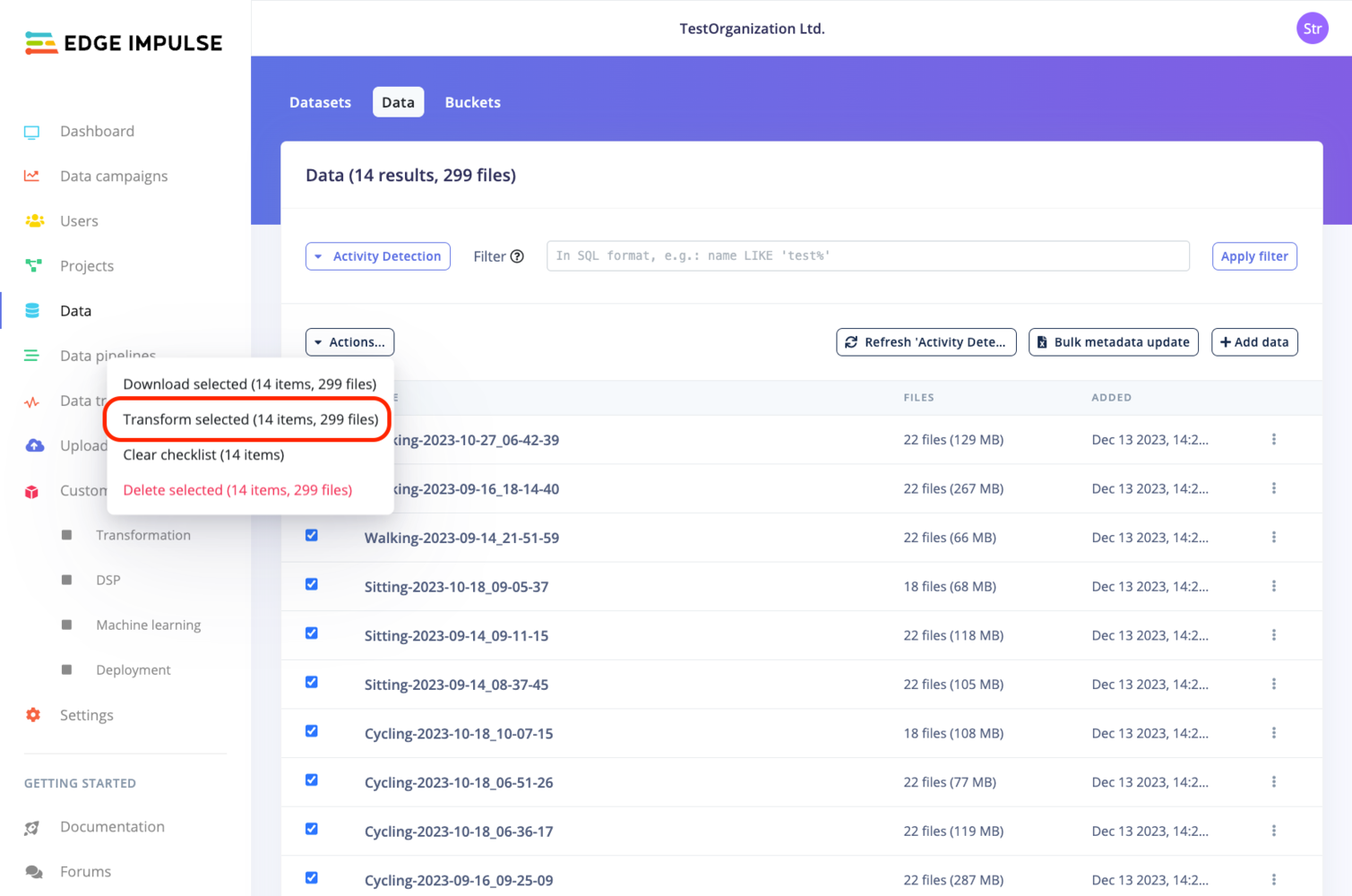
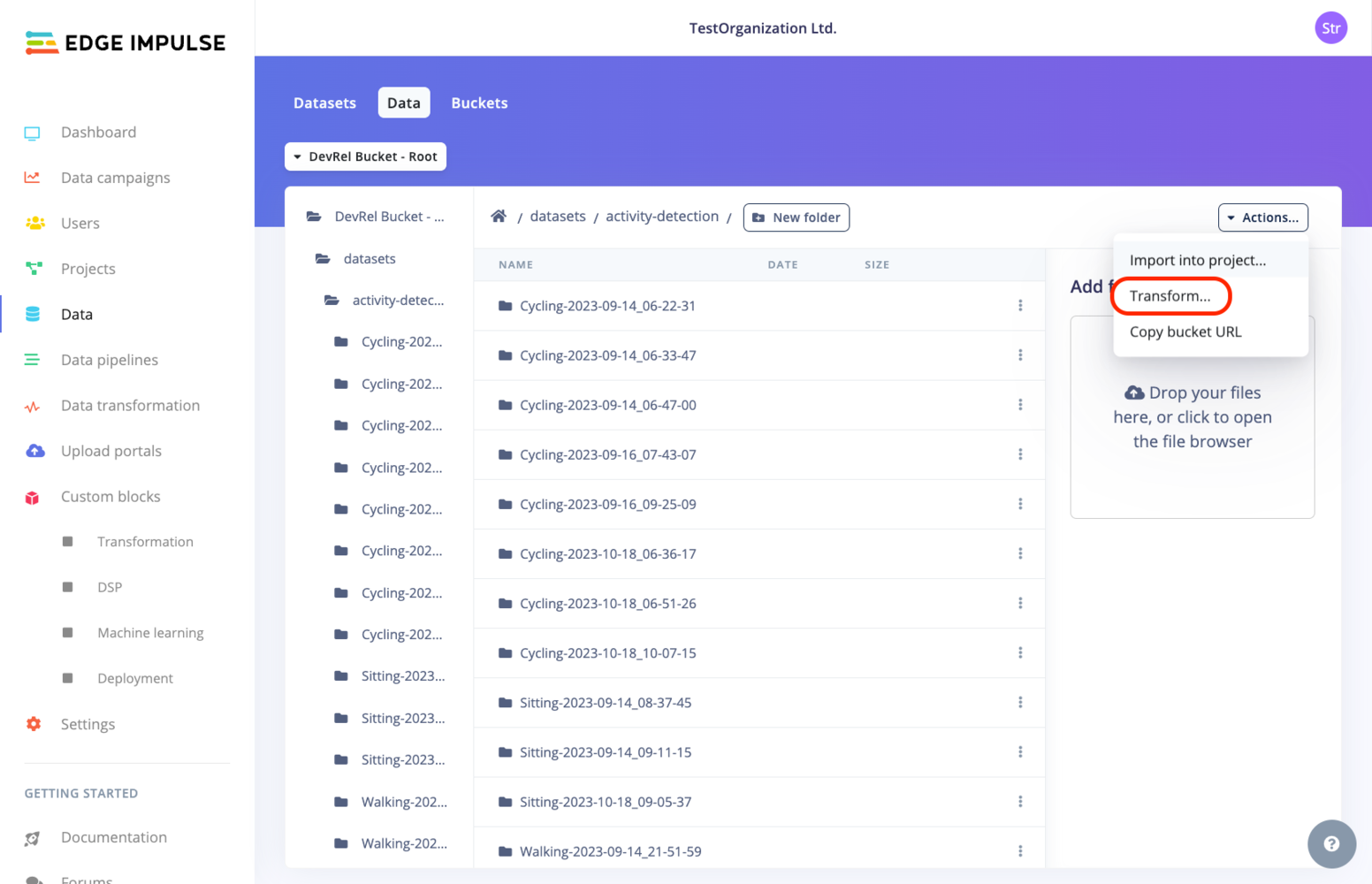
You have several options to create a transformation job:- From the Data transformation page by selecting the Create job tab.
- From the Custom blocks->Transformation page by selecting the ”⋮” action button and selecting Run job.
- From the Data page:
- Default dataset
- Clinical dataset
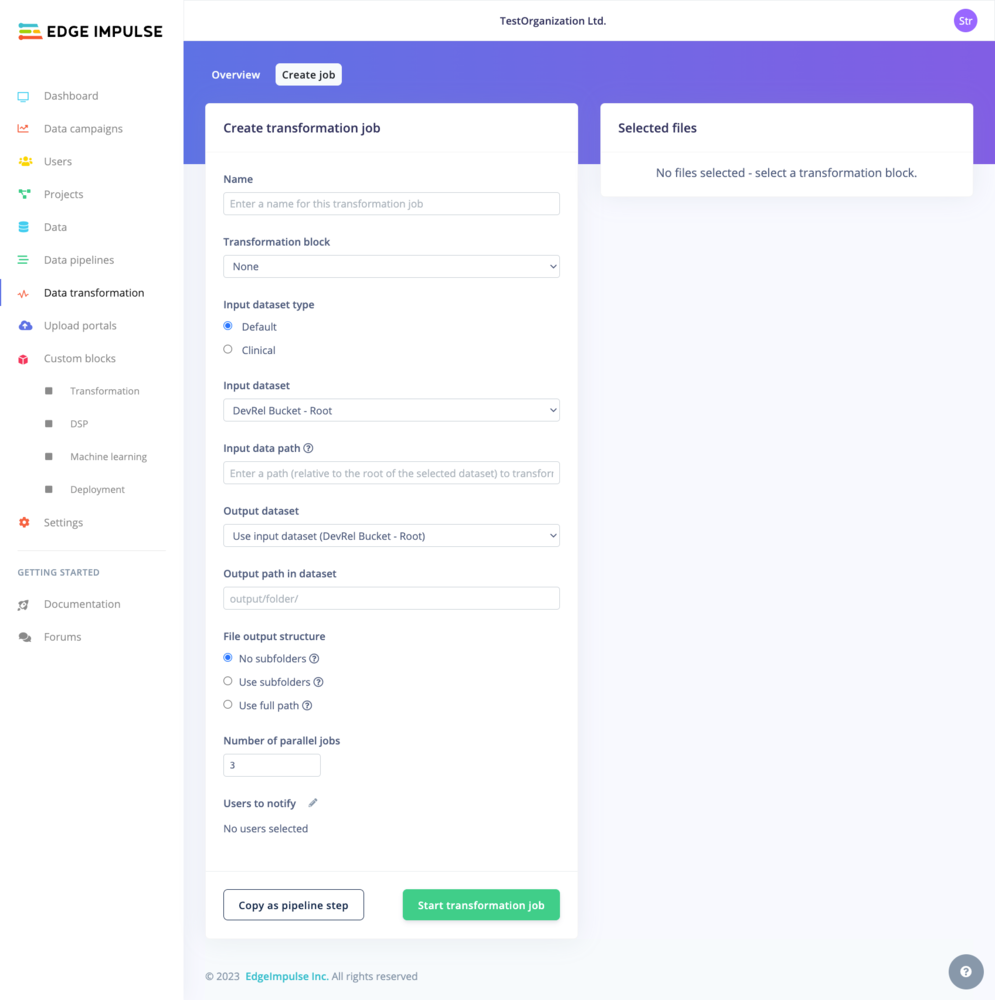
Run a transformation job
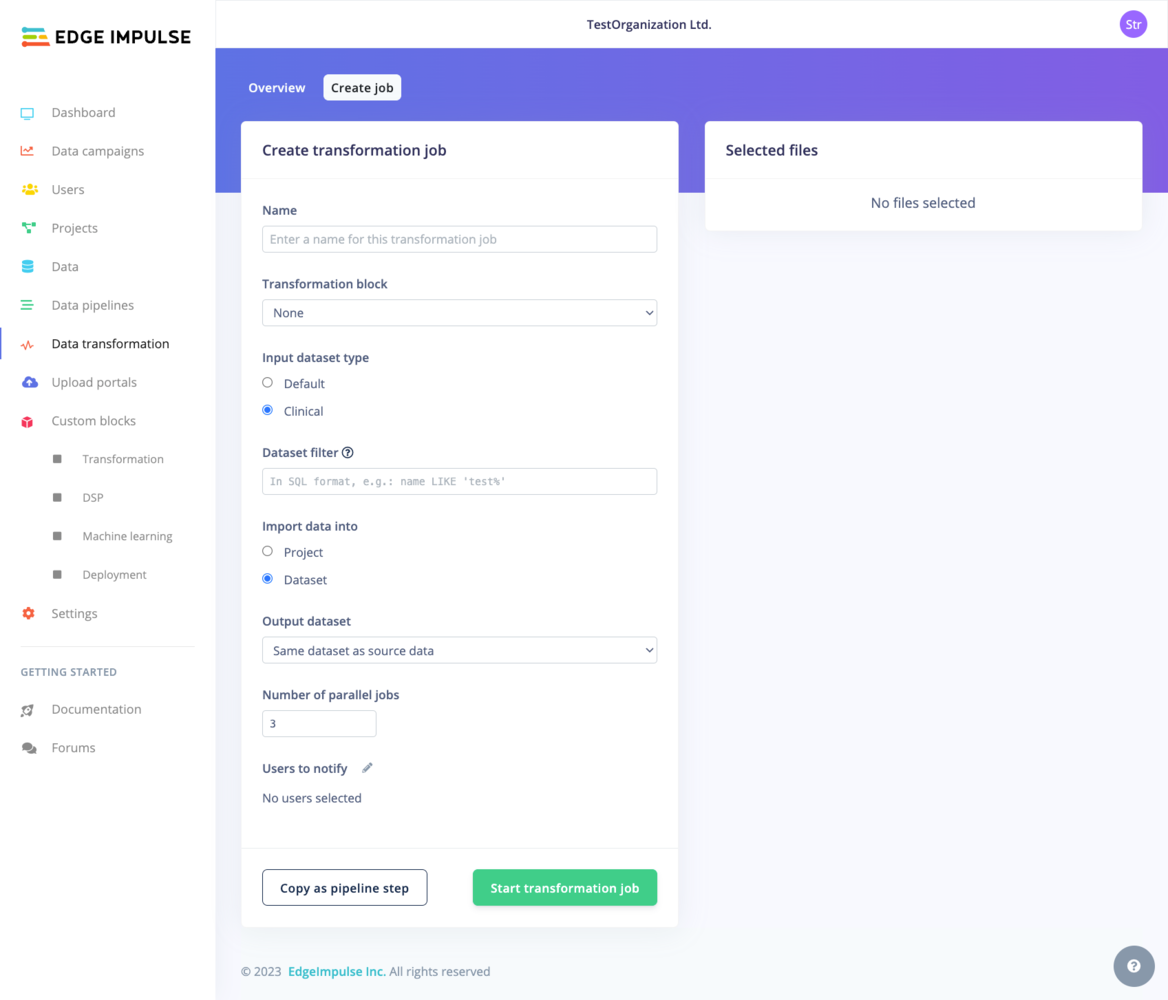
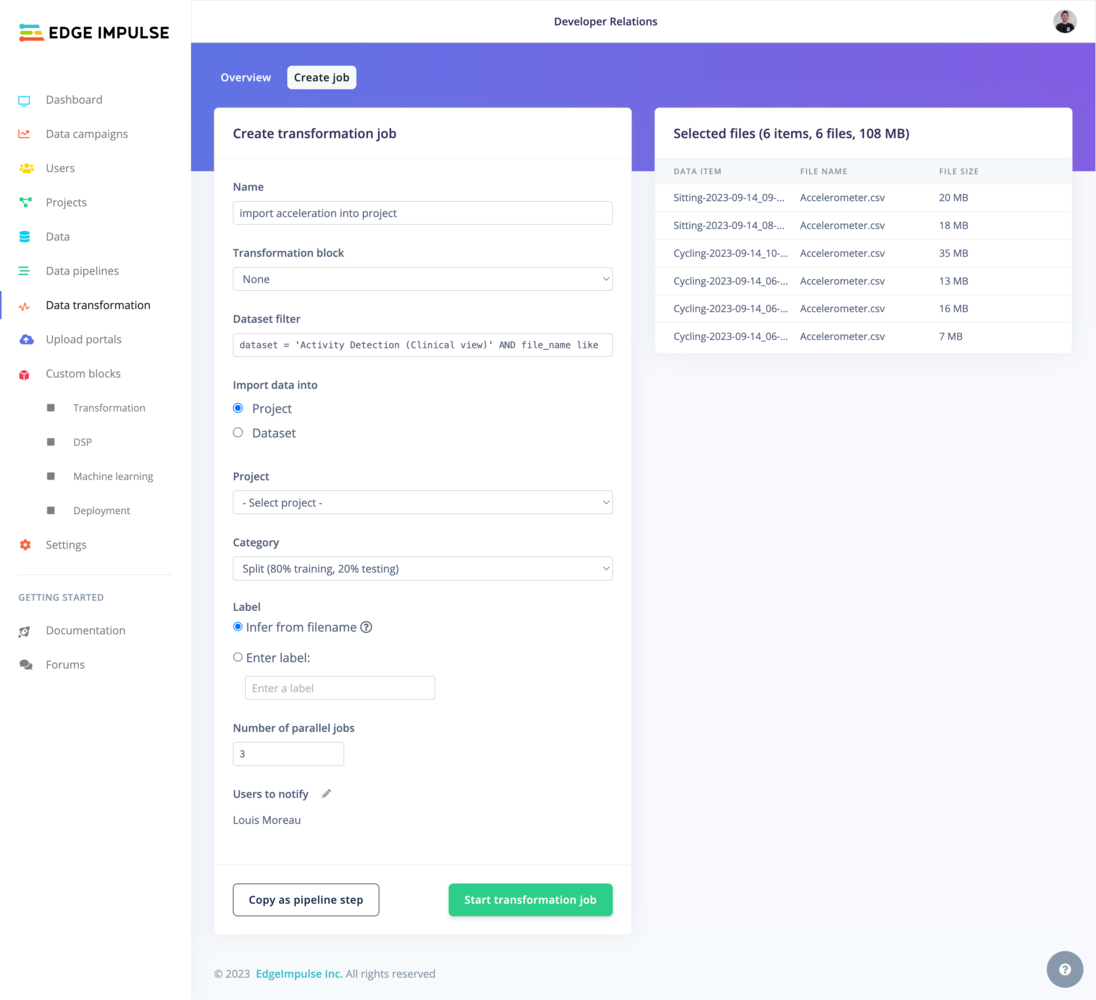
Again, depending on whether you are on a Default dataset or a Clinical dataset, the view will vary. The common options are the Name of the transformation job, the Transformation block used for the job. If your Transformation block has additional custom parameters, the input fields will be displayed below in a Parameters section. For example: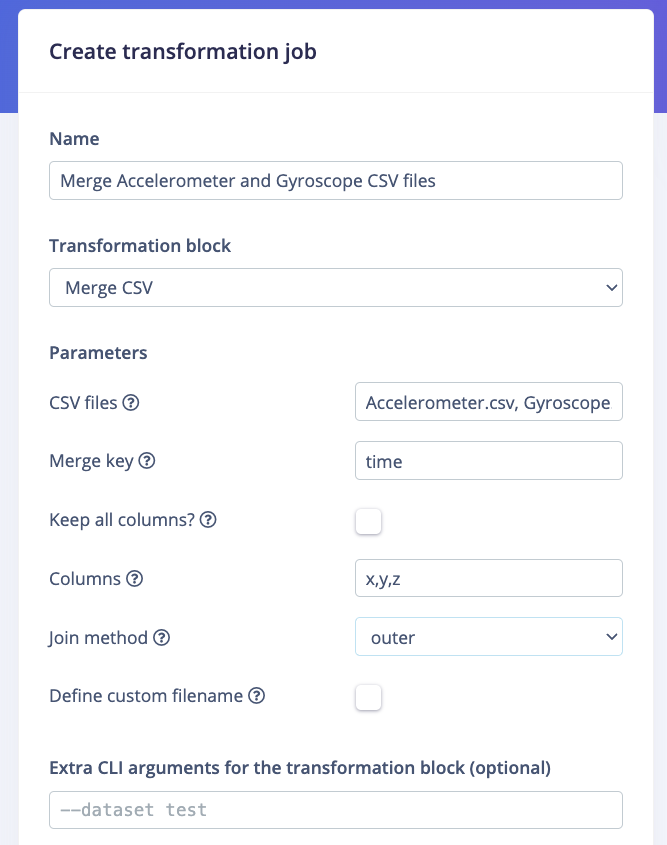
Default vs. Clinical datasets
Clinical Datasets: Operate on “data items” with a strict file structure. Transformation is specified using SQL-like syntax.Default Datasets: Resemble a typical file system with flexible structure. You can specify data for transformation using wildcards.For more information about the two dataset types, see the dedicated Data page.- Default dataset
- Clinical dataset
-
Asterisk ( * ): Represents any number of characters (including zero characters) in a filename or directory name. It is commonly used to match files of a certain type or files whose names follow a pattern.
Example:
/folder/*.pngmatches all PNG files in the/folderdirectory. Example:/data/*/results.csvmatches any results.csv file in a subdirectory under/data. -
Double Asterisk ( ** ): Used to match any number of directories, including nested ones. This is particularly useful when the structure of directories is complex or not uniformly organized.
Example:
/data/**/experiment-*matches all files or directories starting withexperiment-in any subdirectory under/data.
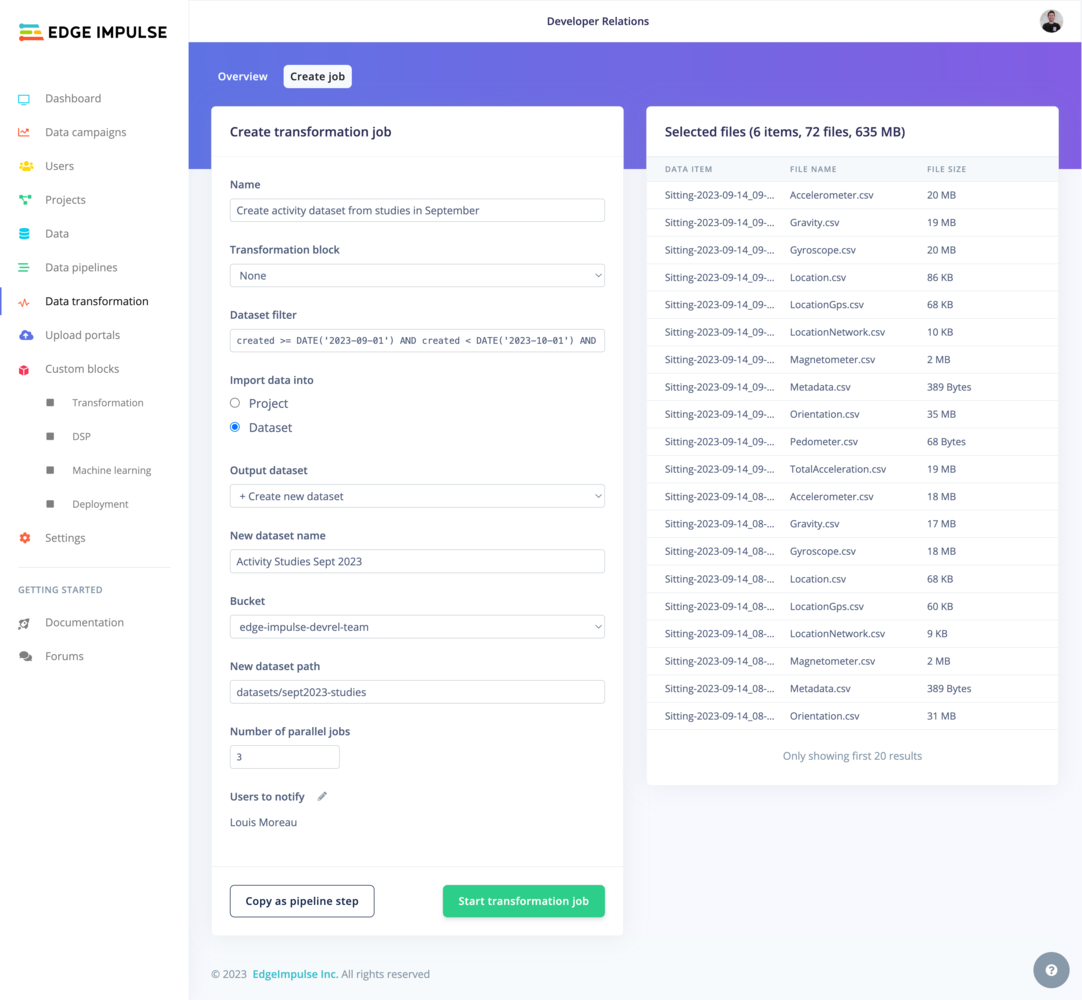
- No Subfolders: This rule places all transformed files directly into your specified output directory, without creating any subfolders. For example, if you transform
.txtfiles in/dataand choose/outputas your output directory, all transformed files will be saved directly in/output. - Subfolder per Input Item: Here, a new subfolder is created in the output directory for each input file or folder. This keeps the output from each item organized and separate. For instance, if your input includes folders like
/data/2020,/data/2021, and/data/2022, and you apply this rule with/transformedas your output directory, you will get subfolders like/transformed/2020,/transformed/2021, and/transformed/2022, each containing the transformed data from the corresponding input year. - Use Full Path: This rule mirrors the entire input path when creating new sub-folders in the output directory. It’s especially useful for maintaining a clear trace of where each piece of output data originated, which is important in complex directory structures. For example, if you’re transforming files in
/project/data/experiments, and you choose/resultsas your output directory, the output will follow the full input path, resulting in transformed data being stored in/results/project/data/experiments.
/activity-detection/Accelerometer.csv will be uploaded to /activity-detection-output/Accelerometer/.