Additional file types or annotation formatsIf none of these above choices are suitable for your project, you can also have a look at custom transformation blocks to parse your data samples to create a dataset supported by Edge Impulse.
Edge Impulse DatasetsNeed inspiration? Check out our Edge Impulse datasets collection that contains publicly available datasets collected, generated or curated by Edge Impulse or its partners.These datasets highlight specific use cases, helping you understand the types of data commonly encountered in projects like object detection, audio classification, and visual anomaly detection.
Upload data
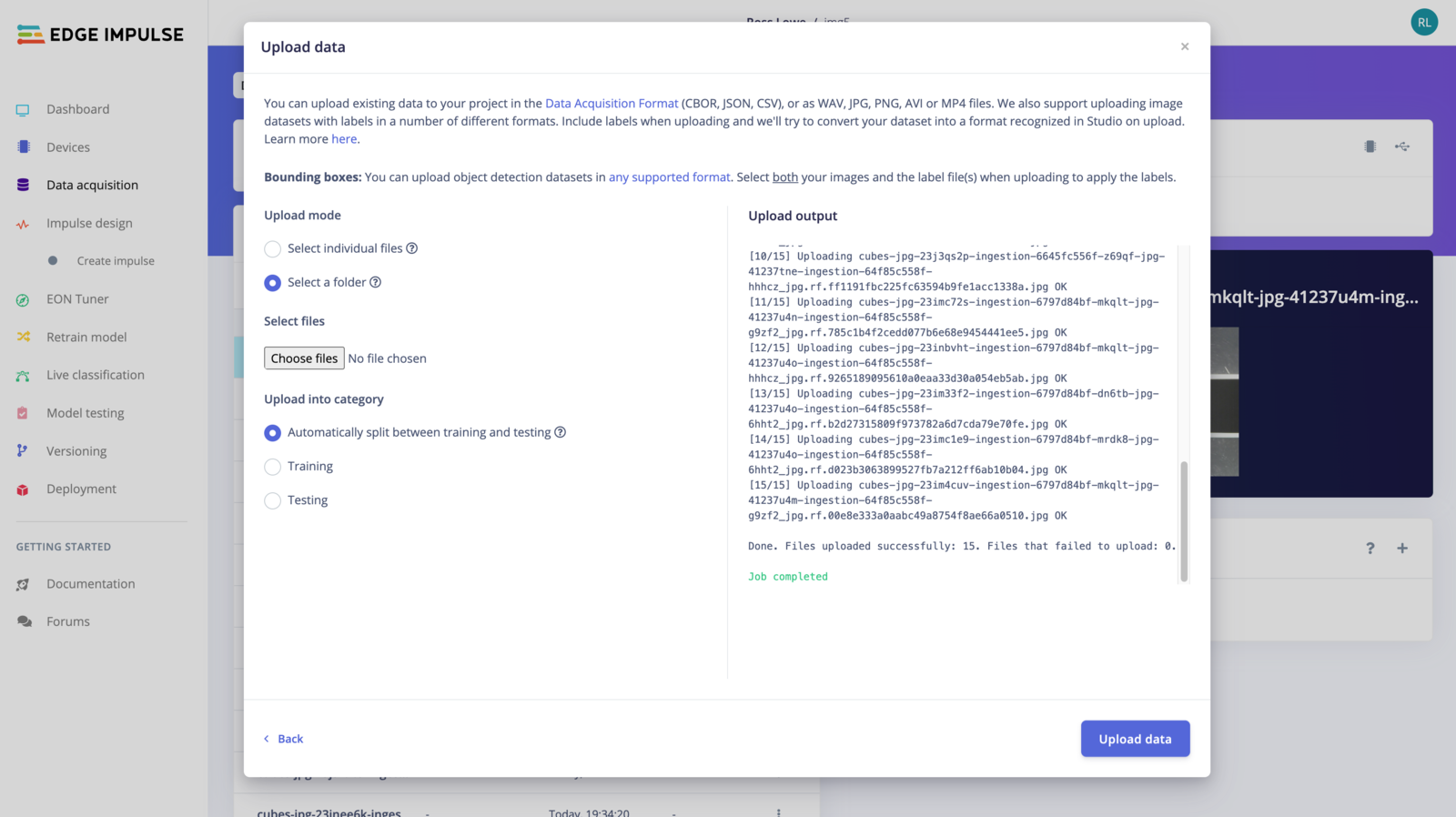
To upload data using the uploader, go to the Data acquisition page and click on the uploader button as shown in the image below: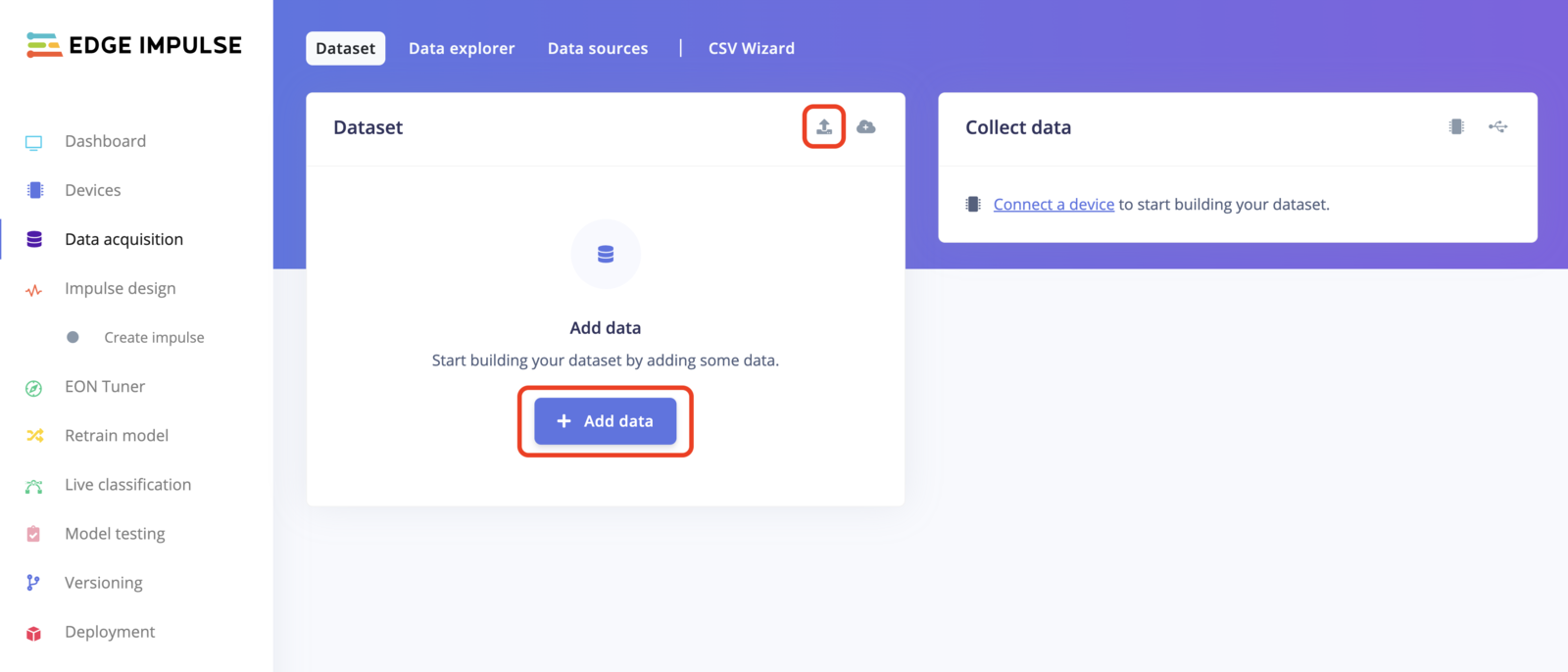
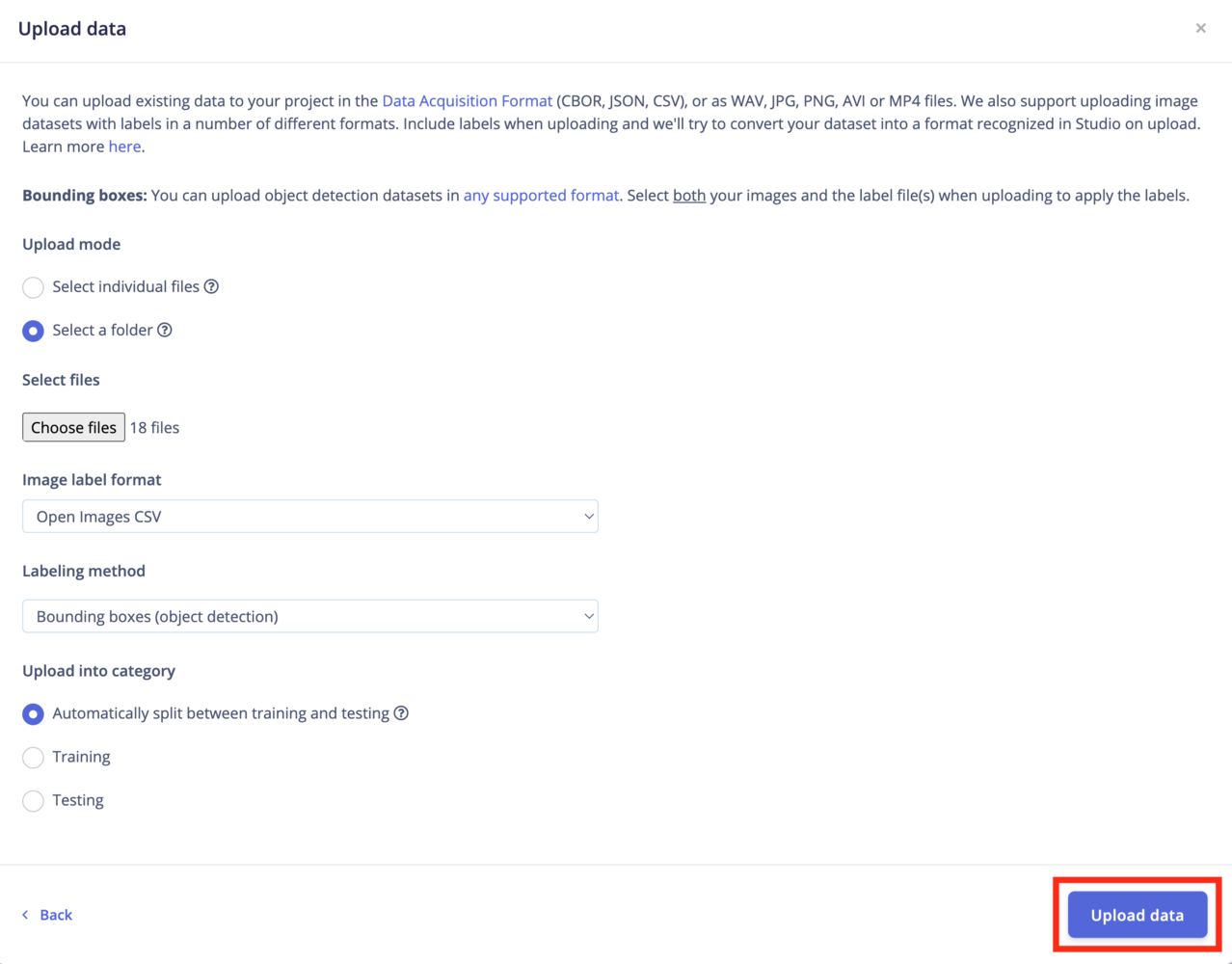
Bounding boxes?If you have existing bounding boxes for your images dataset, make sure your project’s labeling method is set to Bounding Boxes (object detection). You can change this parameter in your project’s dashboard.Then you need to upload any label files with your images. Select both your images and the labels file when uploading to apply the labels. The uploader will try to automatically detect the right format.
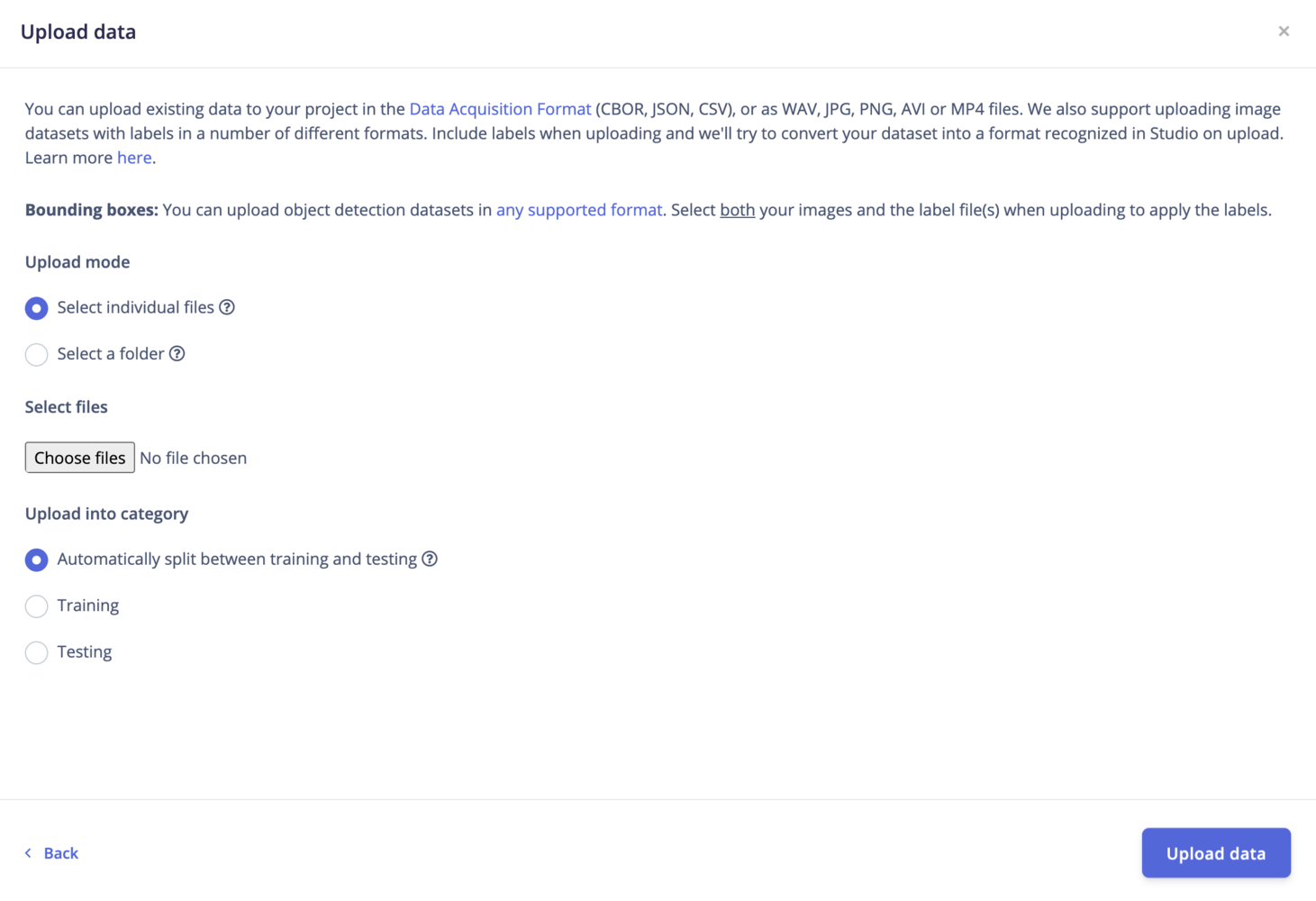
Upload mode
Select individual files: This option let you select multiple individual files within a single folder. If you want to upload images with bounding boxes, make sure to also select the label files. Select a folder: This option let you select one folder, including all the subfolders.Upload into a category
Select which category you want to upload your dataset into. Options can betraining, testing or perform an 80/20 split between your data samples.
If needed, you can always perform a split later from your project’s dashboard.
Label your data
When a labeling method is not provided, the labels are automatically inferred from the filename through the following regex:^[a-zA-Z0-9\s-_]+. For example: idle.01 will yield the label idle.
Thus, if you want to use labels (string values) containing float values (e.g. “0.01”, “5.02”, etc…), automatic labeling won’t work.
To bypass this limitation, you can make an info.labels JSON file containing your dataset files’ info. We also support adding metadata to your samples.
The Studio uploader will automatically detect the info.labels file:
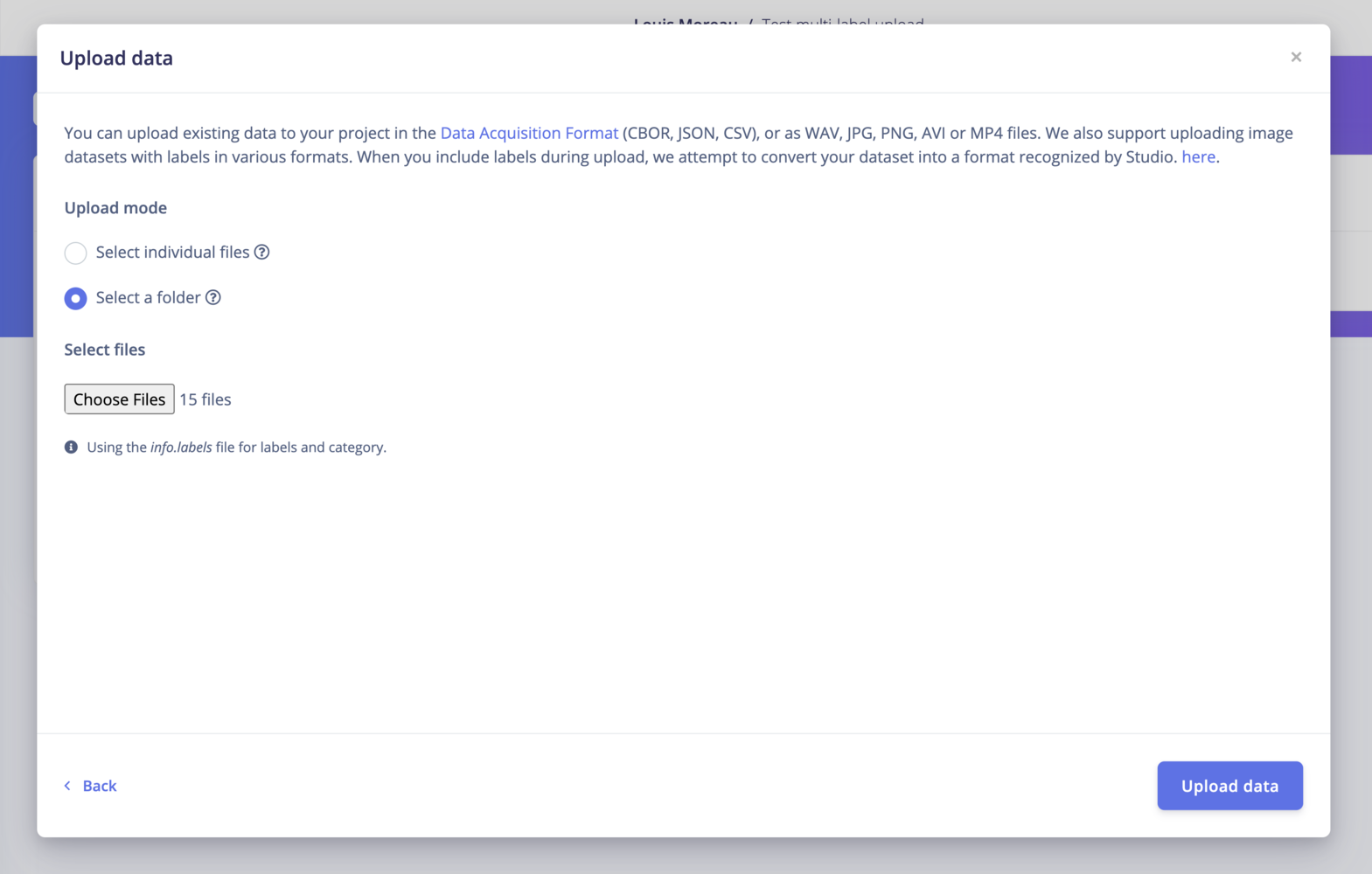
Image dataset annotation formats
Image datasets can be found in a range of different formats. Different formats have different directory structures, and require annotations (or labels) to follow a particular structure. We support uploading data in many different formats in the Edge Impulse Studio.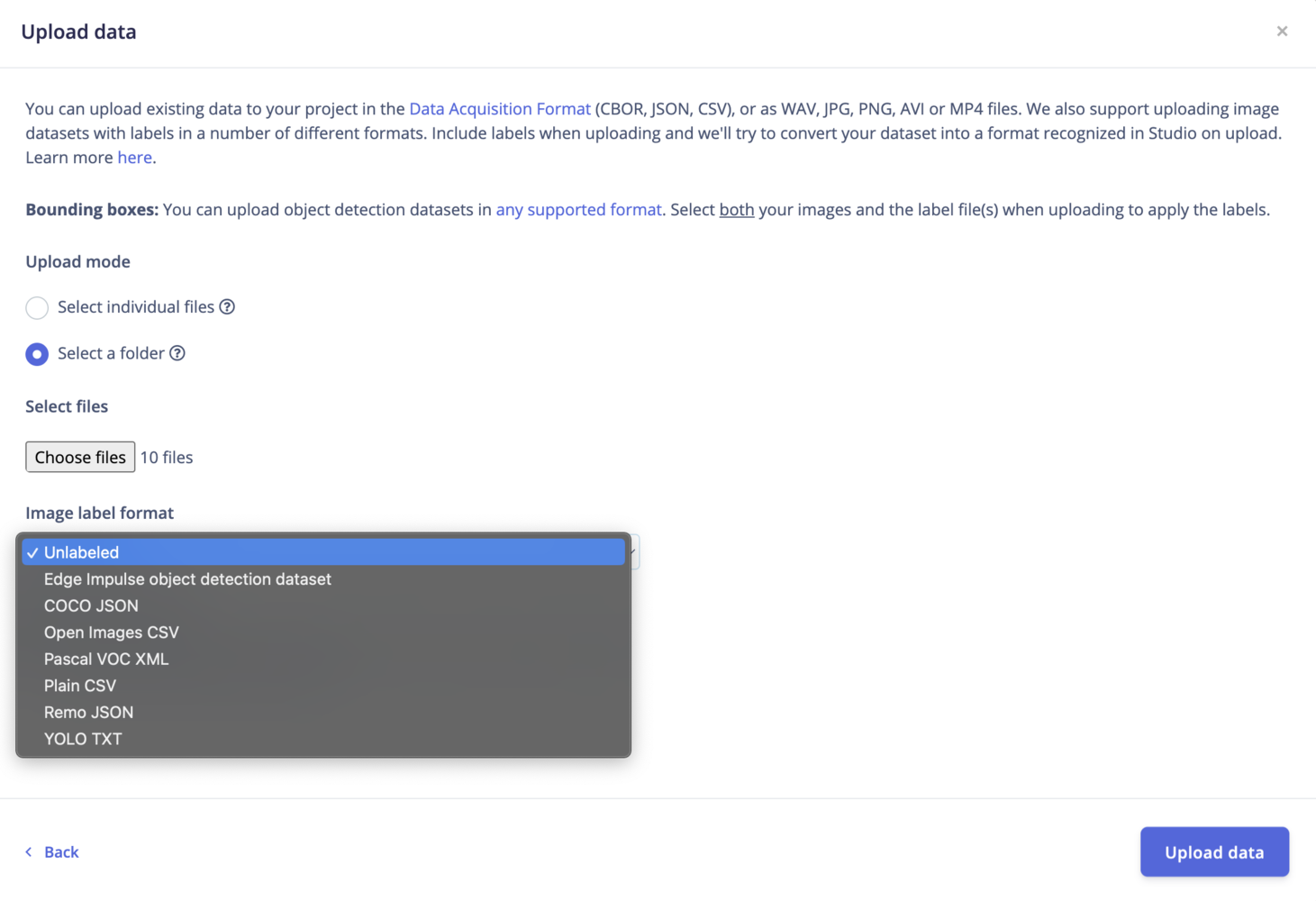
- A single-label: each image has a single label
- Bounding boxes: used for object detection; images contain ‘objects’ to be detected, given as a list of labeled ‘bounding boxes’