Only available on the Enterprise planThis feature is only available on the Enterprise plan. Review our plans and pricing or sign up for our free expert-led trial today.
Block structure
The transformation block structure is shown below. Please see the custom blocks overview page for more details.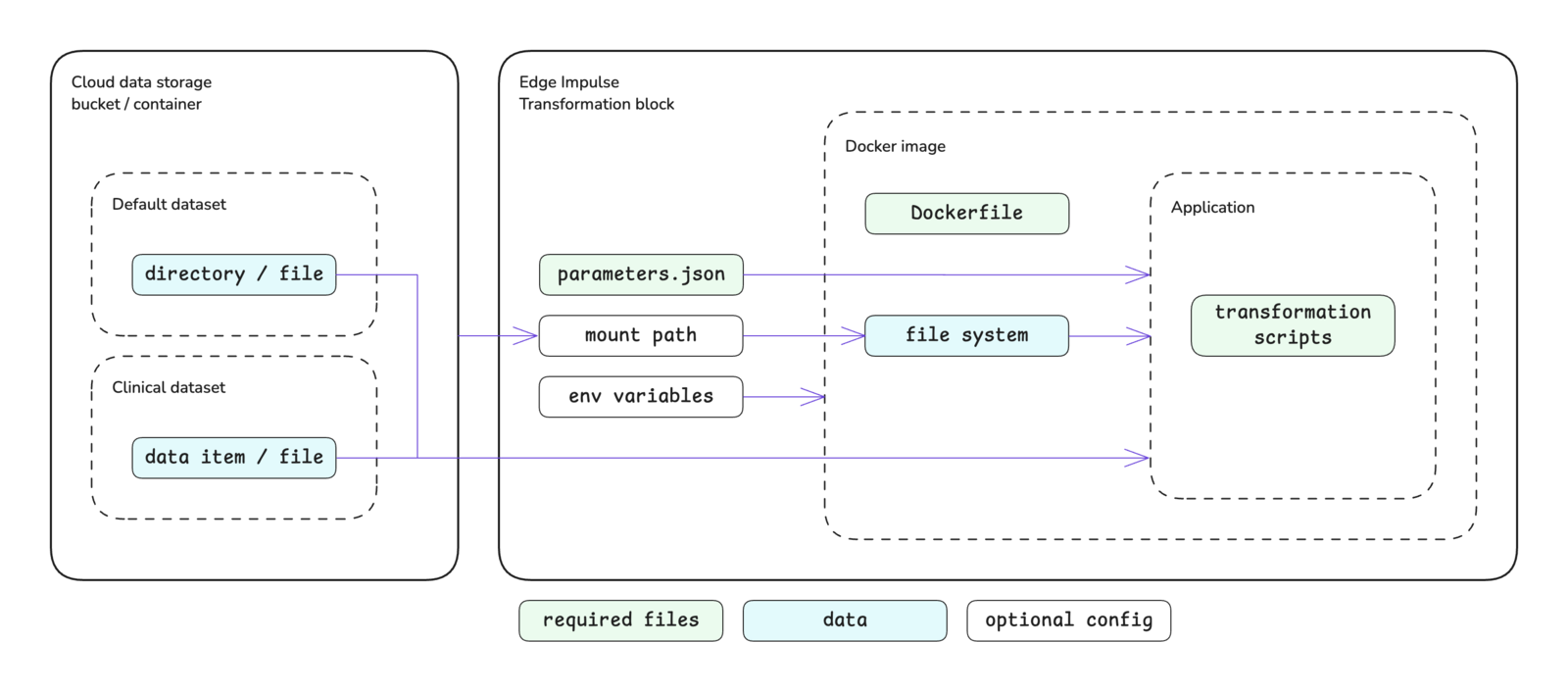
Block interface
The sections below define the required and optional inputs and the expected outputs for custom transformation blocks.Inputs
Transformation blocks have access to environment variables, command line arguments, and mounted storage buckets.Environment variables
The following environment variables are accessible inside of transformation blocks. Environment variable values are always stored as strings.| Variable | Passed | Description |
|---|---|---|
EI_API_ENDPOINT | Always | The API base URL: https://studio.edgeimpulse.com/v1 |
EI_API_KEY | Always | The organization API key with member privileges: ei_2f7f54... |
EI_INGESTION_HOST | Always | The host for the ingestion API: edgeimpulse.com |
EI_LAST_SUCCESSFUL_RUN | Always | The last time the block was successfully run, if a part of a data pipeline: 1970-01-01T00:00:00.000Z |
EI_ORGANIZATION_ID | Always | The ID of the organization that the block belongs to: 123456 |
EI_PROJECT_ID | Conditional | Passed if the transformation block is a data source for a project. The ID of the project: 123456 |
EI_PROJECT_API_KEY | Conditional | Passed if the transformation block is a data source for a project. The project API key: ei_2a1b0e... |
requiredEnvVariables property in the parameters.json file. You will then be prompted for the associated values for these properties when pushing the block to Edge Impulse using the CLI. Alternatively, these values can be added (or changed) by editing the block in Studio after pushing.
Command line arguments
The parameter items defined in yourparameters.json file will be passed as command line arguments to the script you defined in your Dockerfile as the ENTRYPOINT for the Docker image. Please refer to the parameters.json documentation for further details about creating this file, parameter options available, and examples.
In addition to the items defined by you, the following arguments will be automatically passed to your custom transformation block.
| Argument | Passed | Description |
|---|---|---|
--in-file <file> | Conditional | Passed if operation mode is set to file. Provides the file path as a string. This is the file to be processed by the block. |
--in-directory <dir> | Conditional | Passed if operation mode is set to directory. Provides the directory path as a string. This is the directory to be processed by the block. |
--out-directory <dir> | Conditional | Passed if operation mode is set to either file or directory. Provides the directory path to the output directory as a string. This is where block output needs to be written. |
--hmac-key <key> | Conditional | Passed if operation mode is set to either file or directory. Provides a project HMAC key as a string, if it exists, otherwise '0'. |
--metadata <metadata> | Conditional | Passed if operation mode is set to either file or directory, the pass in metadata property (indMetadata) is set to true, and the metadata exists. Provides the metadata associated with data item as a stringified JSON object. |
--upload-category <category> | Conditional | Passed if operation mode is set to file or directory and the transformation job is configured to import the results into a project. Provides the upload category (split, training, or testing) as a string. |
--upload-label <label> | Conditional | Passed if operation mode is set to file or directory and the transformation job is configured to import the results into a project. Provides the upload label as a string. |
cliArguments property in the parameters.json file. Alternatively, these arguments can be added (or changed) by editing the block in Studio.
Lastly a user can be prompted for extra CLI arguments when configuring a transformation job if the allowExtraCliArguments property is set to true.
Mounted storage buckets
One or more cloud data storage buckets can be mounted inside of your block. If storage buckets exist in your organization, you will be prompted to mount the bucket(s) when initializing the block with the Edge Impulse CLI. The default mount point will be:parameters.json file before pushing the block to Edge Impulse or editing the block in Studio after pushing.
Outputs
There are no required outputs from transformation blocks. In general, for blocks operating infile or directory mode, new data is written to the directory given by the --out-directory <dir> argument. For blocks operating in standalone mode, any actions are typically achieved using API calls inside the block itself.
Understanding operating modes
Transformation blocks can operate in one of three modes:file, directory, or standalone.
File
As the name implies, file transformation blocks operate on files. When configuring a transformation job, the user will select a list of files to transform. These files will be individually passed to and processed by the script defined in your transformation block. File transformation blocks can be run in multiple processing jobs in parallel. Each file will be passed to your block using the--in-file <file> argument.
Directory
As the name implies, directory transformation blocks operate on directories. When configuring a transformation job, the user will select a list of directories to transform. These directories will be individually passed to and processed by the script defined in your transformation block. Directory transformation blocks can be run in multiple processing jobs in parallel. Each directory will be passed to your block using the--in-directory <dir> argument.
Standalone
Standalone transformation blocks are a flexible way to run generic cloud jobs that can be used for a wide variety of tasks. In standalone mode, no data is passed into your block. If you need to access your data, you will need to mount your storage bucket(s) into your block. Standalone transformation blocks are run as a single processing job; they cannot be run in multiple processing jobs in parallel.Updating data item metadata
If your custom transformation block is operating indirectory mode and transforming a clinical dataset, you can update the metadata associated with the data item after it is processed.
To do so, your custom transformation block needs to write an ei-metadata.json file to the directory specified in the --out-directory <dir> argument. Please refer to the ei-metadata.json documentation for further details about this file.
Showing the block in Studio
There are two locations within Studio that transformation blocks can be found: transformation jobs and project data sources. Transformation blocks operating infile or directory mode will always been shown as an option in the block dropdown for transformation jobs. They cannot be used as a project data source.
Transformation blocks operating in standalone mode can optionally be shown in the block dropdown for transformation jobs and/or in the block dropdown for project data sources.
For transformation jobs
| Operating mode | Shown in block dropdown |
|---|---|
file | Always |
directory | Always |
standalone | If showInCreateTransformationJob property set to true. |
For project data sources
| Operating mode | Shown in block dropdown |
|---|---|
file | Never |
directory | Never |
standalone | If showInDataSources property set to true. |
Initializing the block
When you are finished developing your block locally, you will want to initialize it. The procedure to initialize your block is described in the custom blocks overview page. Please refer to that documentation for details.Testing the block locally
To speed up your development process, you can test your custom transformation block locally. There are two ways to achieve this. You will need to have Docker installed on your machine for either approach.With blocks runner
For the first method, you can use the CLIedge-impulse-blocks runner tool. See Block runner for additional details.
If your custom transformation block is operating in either file or directory mode, you will be prompted for information to look up and download data (a file or a directory) for the block to operate on when using the blocks runner. This can be achieved by providing either a data item name (clinical data) or the path within a dataset for a file or directory (clinical or default data). You can also specify some of this information using the blocks runner command line arguments.
| Argument | Description |
|---|---|
--dataset <dataset> | Transformation blocks in file or directory mode. Files and directories will be looked up within this dataset. If not provided, you will be prompted for a dataset name. |
--data-item <data-item> | Clinical data only. Transformation blocks in directory mode. The data item will be looked up, downloaded, and passed to the container when it is run. If not provided, you will be prompted for the information required to look up a data item. |
--file <filename> | Clinical data only. Transformation blocks in file mode. Must be used in conjunction with --data-item <data-item>. The file will be looked up, downloaded, and passed to the container when it is run. If not provided, you will be prompted for the information required to look up a file within a data item. |
--skip-download | Skips downloading the data. |
--extra-args <args> | Additional arguments for your script. |
--extra-args <args> argument.
ei-block-data directory within your custom block directory. It will contain subdirectories for the data that has been downloaded.
With Docker
For the second method, you can build the Docker image and run the container directly. You will need to pass any environment variables or command line arguments required by your script to the container when you run it. If your transformation block operates in eitherfile or directory mode, you will also need to create a data/ directory within your custom block directory and place your data used for testing here.
file mode:
directory mode:
standalone mode:
Pushing the block to Edge Impulse
When you have initialized and finished testing your block locally, you will want to push it to Edge Impulse. The procedure to push your block to Edge Impulse is described in the custom blocks overview page. Please refer to that documentation for details.Using the block in Studio
After you have pushed your block to Edge Impulse, it can be used in the same way as any other built-in block.Examples
Edge Impulse has developed several transformation blocks, some of which are built into the platform. The code for these blocks can be found in public repositories under the Edge Impulse GitHub account. The repository names typically follow the convention ofexample-transform-<description>. As such, they can be found by going to the Edge Impulse account and searching the repositories for example-transform.
Note that when using the above search term you will come across synthetic data blocks as well. Please read the repository description to identify if it is for a transformation block or a synthetic data block.
Further, several example transformation blocks have been gathered into a single repository:
Troubleshooting
Files in storage bucket cannot be accessed
Files in storage bucket cannot be accessed
If you cannot access the files in your storage bucket, make sure that the mount point has been properly configured and that you are referencing this location within your processing script.You can double check the mount point by looking at the additional mount point section when editing the block in Studio: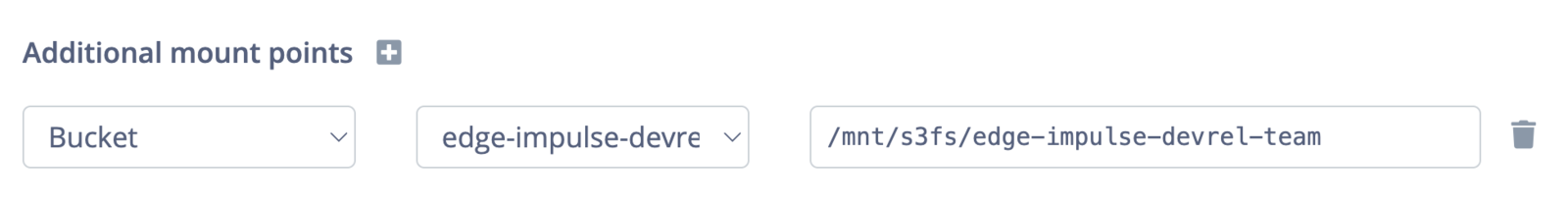

<space> key to select the bucket for mounting (and therefore the storage bucket is not added to the parameters.json file).Transformation job runs indefinitely
Transformation job runs indefinitely
If you notice that your transformation job runs indefinitely, it is probably because of an error with your processing script or the script has not been properly terminated.Make sure to exit your script with code 0 (
return 0, exit(0) or sys.exit(0)) for success or with any other error code for failure.